
Bio
I am Abinash Pant, originally from Kathmandu, Nepal, but I have lived and worked in India, France, Australia and Germany. Currently, I am employed as a Software Engineer at Amazon in Aachen, Germany, since January 2022. My responsibilities include developing and maintaining deep learning toolkits for Automatic Speech Recognition in Alexa. Prior to Amazon, I worked as a Research Scholar and Software Developer at Besa GmbH and the University of Muenster, where I developed automatic segmentation algorithms for MRIs of the head. My work on robust segmentation algorithms has contributed to a better understanding of neurocognitive disorders. By training, I am an electronics and communications engineer with a master in computer science. I completed my Bachelor of Technology from NIT Allahabad, India after receiving a scholarship from the Government of India. I then pursued a Masters in Computer Vision from the Universite de Bourgogne in France. Throughout my academic journey, I have been involved in various internship projects and experiences. My master's thesis focused on Hippocampus Segmentation as an early biomarker for Alzheimer's disease at CSIRO, Australia. I researched the state of the algorithms proposed in the literature for hippocampus segmentation and developed an automated pipeline for hippocampus segmentation using a database of manually segmented hippocampus for training. My areas of interest include Machine Learning, Programming, and Research. In my free time, I enjoy reading, traveling, and solving puzzles.
Education & Experience
-
2022 -Software Developer, Alexa - Amazon, Aachen, Germany
 As a Software Engineer at Amazon in Aachen, Germany I am working towards development and maintenance of deep learning toolkits for Automatic Speech Recognition in Alexa.
As a Software Engineer at Amazon in Aachen, Germany I am working towards development and maintenance of deep learning toolkits for Automatic Speech Recognition in Alexa. -
2015 - 2021Software Developer/Researcher, BESA GmbH, Munich, Germany
 As a Marie Skłodowska-Curie research fellow, I conducted independent research on "Developing automatic segmentation algorithms for MRIs of children." My work also involved research and software development for medical research software, focusing on EEG, MEG, and MRI analysis. I contributed to the entire product life cycle, from design and development to release, ensuring a comprehensive and well-rounded approach to the research and its applications.
As a Marie Skłodowska-Curie research fellow, I conducted independent research on "Developing automatic segmentation algorithms for MRIs of children." My work also involved research and software development for medical research software, focusing on EEG, MEG, and MRI analysis. I contributed to the entire product life cycle, from design and development to release, ensuring a comprehensive and well-rounded approach to the research and its applications. -
2015Intern, CSIRO, Brisnbane, Australia
 Research the state of the algorithms proposed in the literature for hippocampus segmentation Developing an automated pipeline for hippocampus segmentation using a database of manually segmented hippocampus for training.
Research the state of the algorithms proposed in the literature for hippocampus segmentation Developing an automated pipeline for hippocampus segmentation using a database of manually segmented hippocampus for training. -
2014Summer Intern, IRCAD Institute in University Hospital of Strasbourg, France
 Developing a multi-sensor computer vision system to perceive and model clinician and staff activities in an operating room. The project was mainly aimed at estimating human pose for radiation monitoring by using multiple RGB-D cameras to improve results.
Developing a multi-sensor computer vision system to perceive and model clinician and staff activities in an operating room. The project was mainly aimed at estimating human pose for radiation monitoring by using multiple RGB-D cameras to improve results. -
2013 - 2015Master in Computer Vision, Université de Bourgogne, France
 During my master's course at the University of Burgundy, I explored the captivating world of robotics, computer vision, and medical imaging. I mastered cutting-edge techniques, designed innovative solutions, and gained hands-on experience, preparing me for a successful career in these rapidly evolving fields.
During my master's course at the University of Burgundy, I explored the captivating world of robotics, computer vision, and medical imaging. I mastered cutting-edge techniques, designed innovative solutions, and gained hands-on experience, preparing me for a successful career in these rapidly evolving fields. -
2013 - 2011Member of Technical Staff, Spot, Inc., India
 During my tenure, I was involved in building a search and organization platform for personal cloud data, which greatly enhanced user experience and data management. My responsibilities included concept development and rapid prototyping, ensuring that the platform was both innovative and efficient. Furthermore, I contributed to the full product life cycle, from design and development to app store deployment, guaranteeing a seamless and successful launch.
During my tenure, I was involved in building a search and organization platform for personal cloud data, which greatly enhanced user experience and data management. My responsibilities included concept development and rapid prototyping, ensuring that the platform was both innovative and efficient. Furthermore, I contributed to the full product life cycle, from design and development to app store deployment, guaranteeing a seamless and successful launch. -
2008Software Developer, MakeMyTrip.com, India
 From 2010 to 2011, I worked as a Software Developer at MakeMyTrip.com in Gurgaon, India. During my time there, I focused on designing and developing back-end services and web-based solutions related to the travel domain, enhancing the overall user experience. Additionally, I designed and implemented a Finite-state machine solution that streamlined operations, ensuring a more efficient and effective workflow for the company.
From 2010 to 2011, I worked as a Software Developer at MakeMyTrip.com in Gurgaon, India. During my time there, I focused on designing and developing back-end services and web-based solutions related to the travel domain, enhancing the overall user experience. Additionally, I designed and implemented a Finite-state machine solution that streamlined operations, ensuring a more efficient and effective workflow for the company. -
2008Intern, Asian Institute of Technology, Thailand
 During my summer internship at the Asian Institute of Technology (AIT) in Bangkok, I designed and implemented an advanced FPGA-based system using Verilog. I successfully developed a pipelined FFT/IFFT processor utilizing the radix 4 method, that helped me accquire innovative problem-solving skills in the field of digital signal processing.
During my summer internship at the Asian Institute of Technology (AIT) in Bangkok, I designed and implemented an advanced FPGA-based system using Verilog. I successfully developed a pipelined FFT/IFFT processor utilizing the radix 4 method, that helped me accquire innovative problem-solving skills in the field of digital signal processing. -
2006 - 2010Motilal Nehru National Institute Of Technology, B.Tech Electronics & Communications, India
 Electronics & Communications
Electronics & Communications
Research Summary
Electroencephalography (EEG) is a non-invasive and widely used technique to measure electric potential differences that result from current flow in active brain regions. Localization of the relevant brain regions causing the effects on the scalp surface cannot be performed unequivocally due to the so-called “inverse problem”, i.e. different underlying brain activity patterns can cause the same EEG representation. One important factor for identifying the truly active brain regions is having very good knowledge about the so-called “forward model”. As electric current is diffracted differently and reflected to varying degrees by different tissues, it is important to know the anatomy of the head, as well as the conductivity properties of the head tissues as precisely as possible.
Head models representing the conductivity distribution inside the subject's or patient's head play an important role for the solution of the forward, and thus, also the inverse problem. Realistic models taking into account the individual anatomy of the subject can greatly improve the accuracy of source analysis.
Generation of the head model can be accurately estimated with the help of structural imaging techniques. These structural images can be used to segment the different brain regions. An accurate head model could be obtained by using these images to estimate the underlying structure by segmenting the different brain regions. Pipeline to generate head models using segmentation is particularly challenging for children; smaller anatomical structures, motion artifacts, and an anatomy which varies greatly for children at different ages require the development of new algorithms for an accurate and robust segmentation. Development will focus on Bayesian segmentation approaches representing the typical anatomy using Markov Random Field models. Furthermore, age-specific a-priori knowledge on the anatomy of the children's heads will be taken into account.
Interests
- Machine Learning
- Segmentation
- Scene Understanding
- Computer Vision
- Robotics
- Algorithms
Research Projects
-
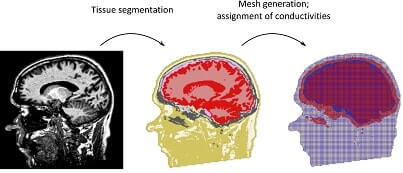
Electroencephalography (EEG) is a non-invasive and widely used technique to measure electric potential differences that result from current flow in active brain regions. Localization of the relevant brain regions causing the effects on the scalp surface cannot be performed unequivocally due to the so-called “inverse problem”, i.e. different underlying brain activity patterns can cause the same EEG representation. One important factor for identifying the truly active brain regions is having very good knowledge about the so-called “forward model”. As electric current is diffracted differently and reflected to varying degrees by different tissues, it is important to know the anatomy of the head, as well as the conductivity properties of the head tissues as precisely as possible.
Head models representing the conductivity distribution inside the subject's or patient's head play an important role for the solution of the forward, and thus, also the inverse problem. Realistic models taking into account the individual anatomy of the subject can greatly improve the accuracy of source analysis.
Generation of the head model can be accurately estimated with the help of structural imaging techniques. These structural images can be used to segment the different brain regions. An accurate head model could be obtained by using these images to estimate the underlying structure by segmenting the different brain regions. Pipeline to generate head models using segmentation is particularly challenging for children; smaller anatomical structures, motion artifacts, and an anatomy which varies greatly for children at different ages require the development of new algorithms for an accurate and robust segmentation. Development will focus on Bayesian segmentation approaches representing the typical anatomy using Markov Random Field models. Furthermore, age-specific a-priori knowledge on the anatomy of the children's heads will be taken into account.
-
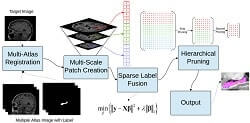
The major objective of this study is to devise an efficient and accurate method for segmenting the hippocampus. The hippocampus being a small part of the brain is quite hard to segment. An accurate segmentation would however make it possible to measure its volume which, in turn, can be used as a biomarker to detect and track the progression of Alzheimer's disease in a patient's brain.
This work provides an extensive study of existing methods for segmenting the hippocampus. Inferences and observations from this perusal of existing methods were used to design a robust and accurate segmentation method. A multi-atlas based method was developed to segment the hippocampus, one patch at a time. This was achieved using a reconstruction algorithm based on sparse representation. Furthermore, a smaller feature representation is used along with hierarchical pruning to include more priori, resulting in an accurate segmentation. The work also presents ways of optimizing the method to make it computationally effective by introducing clever heuristics and leveraging existing approaches.
This method was validated using open source datasets and provided an average Dice score of (L:0.892,R:0.893) using affine registration, which is on par with the state of the art. A thorough validation also proved the effectiveness of the proposed improvements. A study was also undertaken to determine the effectiveness of the method to produce accurate volume. The volume computed using the proposed method was close to the actual volume with an average deviation of 3.87% .
Selected Publications
Filter by type:
Sort by year:
Abstract
Simulated MRI data have great value for segmentation algorithm development since they provide a large set of priors for improved classification accuracy while offering the possibility to introduce imaging anomalies to check and tune the robustness of the algorithm against them. It is possible to achieve this by leveraging a relatively small initial atlas. However, such simulated MRI are not readily available for children’s heads. This along with the prevalence of adult simulated data frequently used as priors (BrainWeb), was our motivation to build an MR simulator for children. The ground-truth and MRI pair obtained from the simulation can be used as priors for segmentation algorithms of complete children’s heads, with the aim of creating realistic head models for EEG/MEG source analysis.
Abstract
Accurate segmentation of an individual human head and the resulting volume conductor model has been known to improve EEG and MEG source analysis. However, comparison of some state-of-the-art segmentation techniques and their effectiveness in source analysis, namely Multi-Atlas and Convolutional Neural Networks (CNN) segmentation, is lacking. We present a comparison between these techniques to segment five tissue types using ground-truth (GT) data from BrainWeb. The comparison of the segmentation techniques shows that 3D CNN outperforms multi-atlas based methods, especially in segmenting WM, GM and CSF. This can be attributed to the structural variability of these three tissue classes across subjects which makes it a challenging problem for segmentation algorithms based on atlas consensus, such as the multi-atlas technique. The CNN's lead-field matrix values were closest to those of the ground-truth which is in accordance with the segmentation results. The effect on source analysis will be investigated further.
An automatic Markov Random Field-based approach for segmentation of volume conductor models of the human head
Biomag 2016 Symposium
Abstract
The accurate solution of the EEG and MEG forward problem requires taking into account information about a subject's individual anatomy. This information is contained in the volume conductor model which describes the electrical properties of the subject's head. Commonly, individual models are constructed by first segmenting the head into the different tissue compartments based on the available medical image data. Next, a suitable discretization of the head domain is computed, and previously published tissue conductivities are assigned. Here, we propose a new automatic segmentation approach for segmenting the tissues of the head. The approach is formulated in a Bayesian framework. A-priori knowledge about the anatomy is included from two sources. A Markov Random Field model encodes our knowledge about the general arrangement of the head tissues. Secondly, a custom probabilistic tissue atlas further facilitates the segmentation. Validation studies versus CT-based and manual segmentations were performed. Results prove the accuracy and reliability of the proposed approach. Average accuracies for the skull segmentation reached values of 88%. In the ChildBrain project, an approach for segmenting volume conductor models of infant subjects and patients will be developed. An outlook will discuss the related challenges and how we are aiming to solve them.
Efficient Multi-scale Patch-Based Segmentation
Conference
Patch-Based Techniques in Medical Imaging
Volume 9467 of the series Lecture Notes in Computer Science pp
205-213
Abstract
The objective of this paper is to devise an efficient and accurate patch-based method for image segmentation. The method presented in this paper builds on the work of Wu et al. with the introduction of a compact multi-scale feature representation and heuristics to speed up the process. A smaller patch representation along with hierarchical pruning allowed the inclusion of more prior knowledge, resulting in a more accurate segmentation. We also propose an intuitive way of optimizing the search strategy to find similar voxel, making the method computationally efficient. An additional approach at improving the speed was explored with the integration of our method with Optimised PatchMatch. The proposed method was validated using the 100 hippocampus images with ground truth segmentation from ADNI-1 (mean DSC = 0.892) and the MICCAI SATA segmentation challenge dataset (mean DSC = 0.8587).
Automated segmentation and T2-mapping of the posterior cruciate ligament from MRI of the knee: Data from the osteoarthritis initiative
Conference 2016 IEEE 13th
International Symposium on Biomedical Imaging (ISBI)
Abstract
Segmentation and quantitative tissue evaluation of the posterior cruciate ligament (PCL) from MRI will facilitate analyses into the morphological and biochemical changes associated with various knee injuries and conditions such as osteoarthritis(OA). In this paper, we validate a multi-scale patch-based method for automated segmentation of the PCL from multiecho spin-echo T2-map MRI of the knee acquired from 26 asymptomatic volunteers. Volume, length and T2-relaxation properties of the PCL were then estimated and validation was performed against manual segmentations of the T2-map images. We apply the method to an MR dataset of 88 patients from the osteoarthritis initiative to investigate differences in T2-properties of the PCL between knees at different stages of OA. A mean Dice's similarity coefficient of 74.4??4.2% was obtained for the PCL segmentation. Moderate and strong correlations were noted between automated and manual volume, length and median T2-values (rV=0.67, rL=0.88, rT2=0.78). Wilcoxon rank-sum tests showed no significant differences in length and median T2-values of the PCL between patients at variable stages of knee OA.
Contact Me
I would be happy to talk to you if you need my assistance in your research or if you just want to hang out with me
- Mobile : +49 176 761 26801
- me *at* abinashpant.com.np
- abinashpant
- linkedin.com/in/abinashpant